
| HUGE |
Gene/Protein Characteristic Table for KIAA0167 |
|
Link to :
Rouge |
GTOP | SWISS-PROT/TrEMBL | GeneCards| RefDIC | |
| Features : DNA sequence | Protein sequence | Expression | Mapping | |
| Product ID : | ORK00439 |
|---|---|
| Accession No. : | D79989 |
| Description : | Centaurin-gamma 1. |
| HUGO Gene Name : | |
| Clone Name : | ha01026 [Vector Info] |
| Flexi ORF Clone : | pF1KSDA0167
 |
| Source : | Myeloblast cell line (KG-1) |
Features of the cloned DNA sequence |
Description | |
|---|---|---|
Length: 3950 bp
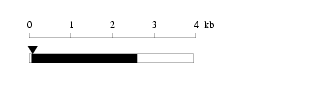
|
| cloned DNA seq. | |
Warning for N-terminal truncation: | NO |
Warning for coding interruption: | NO |
Length of 3'UTR 1351 bp Genome contig ID gi89161190r_56305251 PolyA signal sequence
(ATTAAA,-21)
CTTCTTCAACAGCAATTAAAGAGGAAGTGATTTTGFlanking genome sequence
(100000 - 99951)
TCTAGGGGGTGAGCTGCTGGTCTGTTCTTGGAGGCAGGTCAGGGCATGGC
Chr f/r start end exon identity class ContigView(URL based/DAS) 12 r 56405251 56422209 18 100.0 Perfect prediction
Features of the protein sequence |
Description | |
|---|---|---|
Length: 844 aa
 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.
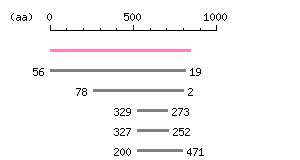 |
|
The numbers on the left and right sides of a black line in the graphical overview indicate the lengths (in amino acid residues) of the non-homologous N-terminal and C-terminal portions flanking the homologous region (indicated by the black line), respectively.

Expression profile |
Description | |
|---|---|---|
RH mapping information |
Description | |
|---|---|---|
| : 12 |
| : Genebridge 4 | |
| : AAACTGGGTGATAGGGCGGAC | |
| : AAGATTCACACCACTGCCTCC | |
| : 215 bp | |
| : 95 °C |
|
How to obtain KIAA clone(s) Back to the HUGE Protein Database homepage 
| |