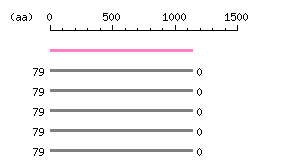
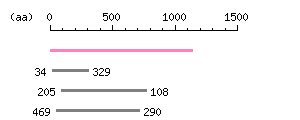
Length: 4051 bp
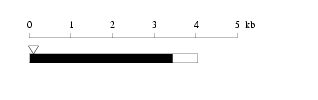
|
| cloned DNA seq. | |
Warning for N-terminal truncation: | NO |
Warning for coding interruption: | NO |
Length of 3'UTR 599 bp Genome contig ID gi89161210r_75974528 PolyA signal sequence
(AATAAA,-23)
ATGTGAACATGTAATAAAATGGAAATGAAATATGGFlanking genome sequence
(100000 - 99951)
AATCCTAAGAACAGAGTTTTACTTATTTGTGTGTGTCTGTGTGTATTGTA
Chr f/r start end exon identity class ContigView(URL based/DAS) 6 r 76074528 76129439 4 100.0 Perfect prediction